Table of Contents
The core of BayesOpt uses standard C/C++ code (C++98) so it can be compiled from many C++ compilers (gcc, clang, MSVC...). The library also include wrappers for Python, Matlab and Octave interfaces.
First, choose the install instructions based on your operating system:
Once you have installed the library with your desired interface, you can Test the installation.
For an advanced installation, you can read Advanced configuration for compilation/install and you can get a detailed explanation of the dependencies needed in Dependencies.
Install in Linux/MacOS
The compilation is very similar in any *nix system. Following these instructions, the library will be compiled using the default configuration. You can modify that easily as explained in Advanced configuration for compilation/install
Dependencies on Linux:
For Ubuntu/Debian, the minimum dependencies (C/C++) can be optained by running:
>> sudo apt install libboost-dev cmake g++
If you want the Python interface or the Octave interface (note that the octave package does not include all the necessary files):
>> sudo apt install python-dev python-numpy >> sudo apt install octave liboctave-dev
You can also install the Python dependencies using your favorite package manager (conda, virtualenv+pip).
You might also want to install the packages for interactive cmake, or GLUT to run the examples with plots:
>> sudo apt install cmake-curses-gui >> sudo apt install freeglut3-dev
And for all dependencies:
>> sudo apt install libboost-dev python-dev python-numpy cmake cmake-curses-gui g++ cython octave liboctave-dev freeglut3-dev
Dependencies on MacOS:
This section assumes homebrew is installed. Similar packages can be found in fink or macports. We also assume that a suitable C++ compiler is installed. For example, Xcode. For the minimal install, run:
>> brew install boost cmake
If you want the Python interface or the Octave interface:
>> brew install python numpy >> brew install octave
You can also install the Python dependencies using your favorite package manager (conda, pip...) or use the native Python interpreter in MacOS. BayesOpt can also be installed in Python environments (conda, virtualenv...).
And for all dependencies:
>> brew install boost python numpy cmake cython octave freeglut
Compile the library with default configuration (C++):
In order to compile the source code in a *nix system, run this from a terminal.
>> cmake . >> make >> sudo make install
Important: If you use ccmake instead of cmake you will access a graphical interface to select features such as the include the Python and Matlab interfaces, debug/release mode or if you want to use shared libraries or not. More details about how to configure it can be found in Advanced configuration for compilation/install
Building the documentation
If you have doxygen installed on your computer, you can compile the documentation right after compiling the code by running.
>> make doc
Thid documentation will appear in the "doc" subdirectory.
Python interface:
Both Python header files (Python.h) and Numpy are needed if you want the python interface. The library has been tested with Python 2 and
- If we want to select the option to compile the Python interface we can just run:
>> cmake -DBAYESOPT_PYTHON_INTERFACE=ON . >> make >> sudo make install
As commented before, the option BAYESOPT_PYTHON_INTERFACE=ON can also be set using
>> ccmake .
You can use and install BayesOpt in a Python environment (conda, virtualenv, pyenv, etc.). It just need to be activated before calling cmake or ccmake. You can check that the correct install path has been found using ccmake. Then, press "t" to select advanced features.
Important: Python requires a special module with shared access and nonstandard name. Thus, it will build a separate module called "bayesopt.so". This module can be accessible from Python provided that it is in the PYTHONPATH or sys.path. It cannot be linked to any executable or other libraries. Use libbayesopt.* instead.
Issues with multiple Python versions:
CMake has a special system to find the Python dependencies. However, sometimes, in systems where there are several instances of Python installed, it can find mixed dependencies.
If you find an error about incorrect Python versions or include files, you can fix this by checking the Python depencies found by CMake. Run:
>> ccmake .
and press "t" to select advanced features. Then, look if the PYTHON_* variables point to the same install/version. Once it has been fixed. Press "c" several times to run the CMake processor. Finally, press "g" to generate the corresponding Makefiles.
MATLAB/Octave interface:
Make sure the library is compiled with the MATLAB_COMPATIBLE option using ccmake. For Mac OS they must be shared.
interface from Octave
Run the script compile_octave.m, which can be found in the /matlab/ directory.
If bayesopt or nlopt are compiled as shared libraries, then, at run time, Octave also needs to access to the libraries. For example, LD_LIBRARY_PATH must include the folder where the libraries are. If the install path is the default, you can execute the exportlocalpath.sh script before calling MATLAB.
interface from Octave
First, configure Matlab to compile mex files if you have not done that before. Run to check the supported compilers and select the appropiate:
>> mex -setup
Run the script compile_matlab.m, which can be found in the /matlab/ directory.
If bayesopt or nlopt are compiled as shared libraries, then, at run time, MATLAB also needs to access to the libraries. For example, LD_LIBRARY_PATH must include the folder where the libraries are. If the install path is the default, you can execute the exportlocalpath.sh script before calling MATLAB.
Issues on libstdc++ linking:
-
Matlab for *nix OS may ship outdated libraries for gcc (e.g.: 4.3 in 2011b). You might get errors like this one:
/usr/lib/x86_64-linux-gnu/gcc/x86_64-linux-gnu/4.5.2/cc1: /usr/local/MATLAB/R2010b/sys/os/glnxa64/libstdc++.so.6: version `GLIBCXX_3.4.14' not found (required by /usr/lib/libppl_c.so.2)
The solution is to change the symbolic links in /matlabroot/sys/os/glnx86 for libgcc_s.so.1 and libstdc++.so.6 to point to the system libraries, which typically can be found in /lib or /usr/lib.
- On MacOS there are known issues both in Matlab and Octave about the compiler linking with the worng std++ library for different reasons. See:
Install in Windows and other systems
Install this components:
- CMake: http://www.cmake.org
- Boost: http://www.boost.org
- A C++ compiler. For example, Visual Studio or MinGW: http://mingw-w64.org/
- FreeGLUT(optional): http://freeglut.sourceforge.net/
First, you need to compile the library from sources using a C++ compiler. If you do not have a C++ compiler, we recomend MinGW+MSYS. In order to configure the compiler and link the dependencies we recomend CMake.
CMake for Windows provides a nice GUI where you can select your favorite C++ compiler (MinGW, Visual Studio, etc.). It will automatically create the necesary configuration files for the compiler (makefile, solution, etc.). In CMake you need to set the paths of the Boost and FreeGLUT to allow the compiler to find them.
Since Boost are pure template libraries, they do not require any install/compilation steps in advance. Just make sure the headers are on the include path. You can also add an entry named BOOST_ROOT in CMake with the corresponding path to the library.
Once CMake have generated the corresponding configuration file, you still need to compile the sources. If you use MinGW, just run:
>> mingw32-make
from the Command Prompt or PowerShell.
If you use Visual Studio, make sure the compilation is set to Release mode and not Debug mode. Although the mode is defined in CMake, Visual Studio tends to ignore that.
The most important options/variables are explained in Advanced configuration for compilation/install.
MATLAB/Octave interface:
Make sure the library is compiled with the MATLAB_COMPATIBLE option and configure Matlab/Octave to compile mex files. For example, in Matlab you can run to check the supported compilers:
>> mex -setup
Run the corresponding script compile_matlab.m or compile_octave.m, which can be found in the /matlab/ directory.
If bayesopt or nlopt are compiled as shared libraries, then, at run time, MATLAB/Octave also needs to access to the libraries. You can modify the PATH variable or copy the dll files in the same bolder as the generated mexfile.
Important: It is strongly recommended to compile bayesopt with exactly the same compiler that was select for mex files. For a list of the supported compilers for your Matlab version, you can check the online docs at mathworks.
MATLAB and MinGW
Although it might not be needed, I have also included a Makefile to generate the mex files outside Matlab. You might need to change the MATLABROOT path with the root folder of your Matlab install and copy the dlls. Then, run mingw32-make.
Python interface:
The Python interface has not been tested in Windows because getting the correct dependencies is highly involved. You might need to download and install:
- Python (binary and headers): http://www.python.org
- Numpy: http://new.scipy.org/download.html
Also, read this article about how to link everything: http://docs.python.org/2/extending/windows.html#building-on-windows
Test the installation.
The library includes several test program that can be found in the bin folder. Furthermore, there are test programs respectively under the python and matlab folders. They provide examples of the different interfaces that BayesOpt provide.
-
Run bo_branin. It should found one of the minima shown at the end of the run.
If this does not work, it might be because the program is not finding the correct dynamic libraries:
-
In Windows, you can copy the DLLs to your working folder or include the lib folder in the system PATH.
-
In some *nix systems, including Ubuntu, Debian and Mac OS, the library is installed by default in /usr/local/lib/. However, this folder is not included by default in the linker or Matlab paths by default. This is specially critical when building shared libraries. The script exportlocalpaths.sh makes sure that the folder with the libraries is included in all the necessary paths.
-
The Python package is compiled in a different file and installed in the proper folder by default (site-packages). Thus, the Python interface should work out of the box in Linux and Mac OS.
-
-
Python: the scrip demo_quad gives a quick overview of all the features of the Python interface. The function to optimize is a 5-dimensional quadratic function
![$x^* = [0.3]^5$](form_11.png) and
and 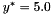 . The discrete version just consider only 100 points randomly distributed in the
. The discrete version just consider only 100 points randomly distributed in the ![$[0,1]^5$](form_13.png) box, so the result might differ from the actual optimum.
box, so the result might differ from the actual optimum. - Matlab/Octave: the scrip runtest also is configured to run different sets of test, the first one also on the Branin function.
Advanced configuration for compilation/install
CMake allows to configure the compilation using some variables (see for example how to compile the Python module in Linux). These variables can be set in Linux/MacOS from the command line with the -D flag:
>> cmake -DVARIABLE=VALUE .
For example
>> cmake -DCMAKE_BUILD_TYPE=Debug .
If you use ccmake in Linux/MacOS or CMake for Windows, you can access to a list of all the variables and their values. Just modify the value of the desired variable.
Compile as shared libraries
We can select if we want BayesOpt and NLOPT compiled as shared libraries
BAYESOPT_BUILD_SHARED=ON NLOPT_BUILD_SHARED=ON
In this case, we also need to force rebuild NLOPT (by default it is not compiled if it is found in the system).
Install the library in a different path
CMake allows to select the install path before compilation compilation. You just need to change the CMAKE_INSTALL_PREFIX variable.
CMAKE_INSTALL_PREFIX=/your/desired/path
Minimal installation (fast compilation)
Sobol sequences can be used to for the initial design (see Initialization parameters). In many cases, the performance is similar to latin hypercube sampling, however including the Sobol components increases considerably the library size and the compilation time. Thus, it can be removed from compilation:
BAYESOPT_BUILD_SOBOL=OFF
Similarly, we can avoid to compile the example files and demos:
BAYESOPT_BUILD_EXAMPLES=OFF
Dependencies
CMake: The easiest way to compile this library is using the cross platform and cross compiler tool CMake.
Boost: This code uses Boost libraries for matrix operations (uBlas), random number generation, math functions and smart pointers. Boost can be found in many Linux and MacOS repositories. It can also be downloaded from http://www.boost.org.
OpenGL/FreeGLUT (optional): It is used for the demos that include visualization.
Python/Numpy (optional): Both Python development files (Python.h) and Numpy are needed if you want the Python interface. The library has been tested with Python 2.6 and 2.7. The interface relies on Numpy arrays.
Matlab/Octave (optional): The library is compatible with any Matlab from 2010 and Octave. If you want the Matlab interface, just make sure you have a C++ compiler compatible with your Matlab version.
NLOPT (included): A LGPL library for nonlinear optimization. It has been slightly modified and include the corresponding CMake files.
Sobol (included): A LGPL library to generate Sobol sequences.
Matplotpp (included): A GPL library for visualization and plotting. The code included in this package includes some bug-fixes and compiling issues. It requires OpenGL/GLUT.
Compatibility tests
| API | Linux | MacOS | Win/MinGW | Win/MSVC |
|---|---|---|---|---|
| C/C++ | OK | OK | OK | OK |
| Matlab | OK | OK | OK | OK |
| Octave | OK | OK | No test | No test |
| Python2/3 | OK | OK | No test | No test |
